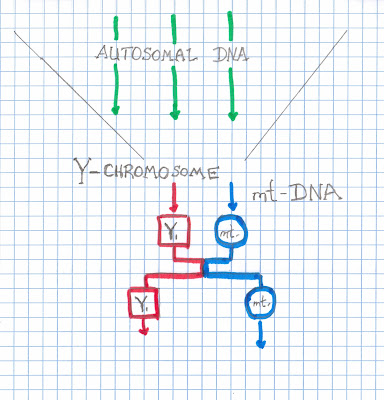
So there you have it. How to understand the origins of all this DNA and genealogy stuff. At least where the terms are derived among the branches. Let's summarize.
1) The first and earliest DNA to be associated with family trees (doing genealogy) comes through the maternal side...known as mtDNA [name from the mitochondrial structures located out side of the nucleus]. This form of DNA gave rise to a nomenclature called "haplogroups" which were associated with geographic locations.
2) Male descent [Y-chromosome] became the next wave of "genetic" genealogy. As the linear sequence of the DNA contained came to be understood, all labs needed to agree upon the labeling. Broken down by the type of changes that have occurred along this linear arrangement, another series of "haplogroups", divided even further to "haplotypes", became the rave of genealogy. The term MRCA [most recent common ancestor ] was used to calculate a likelihood of sharing a common ancestor based upon the matches shared among folks who had this method of DNA tested.
3) The next phase of genetic genealogy expanded the population base [both male and female] that could be tested. Using techniques that "stained" the surface of the 22 chromosomes called "autosomes", this process gave a map of the staining patterns found associated with the various types of specialized proteins called "histones". This form of testing allowed identification of the geographic locations which clustered these changes. Then, the various markers identified among these autosomes could be assigned to the ethic groups they represented, resulting in the "pie" figure now frequently shown.
A simple diagram follows:
The Jones Surname DNA
Connecting loose ends.
Wednesday, May 24, 2017
Tuesday, May 2, 2017
Foundational Concepts of DNA (3)
The various cells of the human body are assigned particular tasks. Stomach cells produce acid, kidney cells save water, and liver cells remove certain items from the blood are all examples. Now while each cell is carrying out its assigned function, the DNA contained in its nucleus lives in a very poorly defined form called "chromatin". Special proteins (many provide structural and enzymatic support) are called "histones", and bind to certain areas along the DNA. When the signal to divide and produce new cells is given, this chromatin, along with its histones, begins a cycle of changes becoming a more visible and distinct form called "chromosomes". Except for the ova/sperm cells, there are 22 pairs of chromosomes in each cell called autosomal. These none sexual cells, called "autosomes" , provide the stages for what is called "autosomal DNA". It is the associated histones (to each chromosome) that are marked and analyzed. This analysis becomes the bases for the reports generated.
To give a sense of the relative size of a chromosome, the follow is my attempt to draw in proportion #1 chromosome [an autosome] in relationship to the X and Y chromosomes [the sex chromosomes].
Foundational concept 3 = chromosomes package our DNA including the associated specialized helper proteins called histones.
To give a sense of the relative size of a chromosome, the follow is my attempt to draw in proportion #1 chromosome [an autosome] in relationship to the X and Y chromosomes [the sex chromosomes].
Foundational concept 3 = chromosomes package our DNA including the associated specialized helper proteins called histones.
Monday, April 3, 2017
Foundational Concepts of DNA (2)
Corporate headquarters for your DNA is the nucleus. At the same time, multiple branch offices reside scattered around that part of the cell out side of the nucleus called the "cytoplasm". These branch offices are called "mitochondria" and contain a much smaller, circular shaped DNA passed down from the moms of the world. You have come to call this mtDNA [ for this unique form of circular DNA located in the branch offices]. A big picture of this single cell organization is shown.
The yellow area is the nucleus. The clear moist looking area is the cytoplasm. Within each branch office [mitochondria] are specialized copy machines called "ribosomes". Communication between the home office and all the branches use special messengers to carryout the purpose of this organization called RNA [ribonucleic acid]. Three RNA forms exist called messenger RNA... transfer RNA... and ribosomal RNA. Universally accepted as the central tenant, the purpose of DNA is to make proteins: 1) new DNA is copied from the existing DNA, 2) messenger RNA is copied from DNA, and 3) proteins are made from this messenger RNA. Wow...what a deal.
The yellow area is the nucleus. The clear moist looking area is the cytoplasm. Within each branch office [mitochondria] are specialized copy machines called "ribosomes". Communication between the home office and all the branches use special messengers to carryout the purpose of this organization called RNA [ribonucleic acid]. Three RNA forms exist called messenger RNA... transfer RNA... and ribosomal RNA. Universally accepted as the central tenant, the purpose of DNA is to make proteins: 1) new DNA is copied from the existing DNA, 2) messenger RNA is copied from DNA, and 3) proteins are made from this messenger RNA. Wow...what a deal.
Monday, March 13, 2017
Foundational Concepts of DNA
The DNA revolution has produced a number of new opportunities for the eager genealogist. Along with these opportunities come a host of unusual terms, fancy words, and confusing outcomes that can make for some, a very difficult experience. For those, who like me, continually express...say what!...this post will begin perhaps a series of posts on some basic ideas surrounding all this DNA stuff. So, let's begin.
Our human existence is dependent on the cell, the basic unit of life. All "us humans" are made up of these cells, and come from cells. As we know it, one of the most important aspect of these cells is reproduction. [Good thing or we would'nt be here!] At any rate, it is during this reproduction that our genetic traits get pass down. Now this DNA ( doxyribonucleic acid ) is the molecule of inheritance for all. These poor cell's DNA is the thing that gets knocked around in all these labs...scraped, dipped, broken apart, multiplied, electrified, analyzed, and...the results printed out, as the "Holy Grail" of our long lost relatives still among the family tree.
The basis for most traits past down is the functional unit of inheritance call the "gene". Each gene is given a specific position (location or loci) along the DNA molecule to be preserved and transmitted on down the family tree. These genes control our biological processes through the production of proteins and a special modification called RNA ( ribonucleic acid ). The genetic composition of all genes is called the "genotype". The observable inherited traits [ physical, behavioral, physiological, etc.] is given the word "phenotype".
Foundational concepts 1) Genes form the basis of our inheritance.
Our human existence is dependent on the cell, the basic unit of life. All "us humans" are made up of these cells, and come from cells. As we know it, one of the most important aspect of these cells is reproduction. [Good thing or we would'nt be here!] At any rate, it is during this reproduction that our genetic traits get pass down. Now this DNA ( doxyribonucleic acid ) is the molecule of inheritance for all. These poor cell's DNA is the thing that gets knocked around in all these labs...scraped, dipped, broken apart, multiplied, electrified, analyzed, and...the results printed out, as the "Holy Grail" of our long lost relatives still among the family tree.
The basis for most traits past down is the functional unit of inheritance call the "gene". Each gene is given a specific position (location or loci) along the DNA molecule to be preserved and transmitted on down the family tree. These genes control our biological processes through the production of proteins and a special modification called RNA ( ribonucleic acid ). The genetic composition of all genes is called the "genotype". The observable inherited traits [ physical, behavioral, physiological, etc.] is given the word "phenotype".
Foundational concepts 1) Genes form the basis of our inheritance.
Monday, August 15, 2016
Haplogroup Hangouts
Our human Y-chromosome has experienced a number of mutations since the dawn of man. On the most part, these mutations have been identified and given a standard nomenclature. Called "haplogroups" , they have been organized by number [DYS #] and alphabetized A - T. Their chronology has been estimated, with "haplogroup A" starting things off some 60,000 years ago, or so. Their geographic locations have been identified. The following figure was drawn some six years ago when trying to put all this DNA stuff together. I thought it might be of interest in the very broadest sense.
The haplogroups are shown in a chronological sense top to bottom. The DYS # associated with each haplogroup is listed. The general "hangout" for each haplogroup(s) is written underneath each. The green swipe represents Africa with migration "out of Africa" identified. Various colors then mark in the broadest terms, the migration patterns thought most likely for each haplogroup. The JONES surname haplogroup is approximaly 75% R1b, and is shown by the darker blue line.
Give it a go. If you see any errors or need for more recent information please comment. Here you have my take on the haplogroup hangouts.
The haplogroups are shown in a chronological sense top to bottom. The DYS # associated with each haplogroup is listed. The general "hangout" for each haplogroup(s) is written underneath each. The green swipe represents Africa with migration "out of Africa" identified. Various colors then mark in the broadest terms, the migration patterns thought most likely for each haplogroup. The JONES surname haplogroup is approximaly 75% R1b, and is shown by the darker blue line.
Give it a go. If you see any errors or need for more recent information please comment. Here you have my take on the haplogroup hangouts.
Wednesday, July 13, 2016
That Y-chromosome (part 4) Other Surnames
The particular haplotype R1b1a2 is one of the most common to carry the surname JONES. It is also defined as R-M269 which is the genetic marker that is tagged for this group. The following chart shows additional surnames that have been found to have the same marker [R-M269] from those who have tested their Y-DNA. The listed is given in alphabetical order for the 61 other surnames that are found to match those with the surname JONES among my own DNA study groups. What a deal!
When testing your own Y-DNA for your JONES surname, you may find that those among this list are more likely to match you than another JONES!
When testing your own Y-DNA for your JONES surname, you may find that those among this list are more likely to match you than another JONES!
Wednesday, March 2, 2016
That Y-chromosome (part 3)
Since our Y-chromosome found its way out of Africa, a number of pathways were followed. The Mediterranean Sea placed a coast line which provided a road to split that Y-chromosome east to west. The northwestern coastal group soon ran into other geographic boundary's with the Black Sea being northwest, and Caspian Sea being northeast, and the Caucasus Mountains in between. It was around this 550 mile mountainous system that our R-haplogroup is thought to have first appeared. I suspect that the ice curtain that kept going up and down also had something to do with the next migration groups. The R1-haplogroup scattered about leaving this Y-chromosome as far northeast as the slopes of the S. Ural mountains [Bashkirs] and the Basque area of western Europe.
Central Europe seemed to be one place that the R1b - haplogroup settled around those salt mines that were so important for early human survival. For our JONES surname things [genetically] took roots both in culture and language. Expanding their horizons they moved about the Iberian peninsula [and other places] and finally found their way up St. George's channel to place their Y-chromosome among the islands. For R1b1a - haplogroup [roughly 75% of us with the JONES surname ] the Welsh (Anglesey) at 89%, and Basque area (French, Spanish) at 88%, and the Turkic people (Bashkirs) were found with 86%. [confines of S.Ural mountains]
For those interested: 1) Irish = 82% , 2) Scots = 77%, 3) Spanish (Minorca) = 73% , 4) Dutch (Germanic west) = 70%. What a deal...that same Y-chromosome is found among most of those with the JONES surname of Welsh descent.
Central Europe seemed to be one place that the R1b - haplogroup settled around those salt mines that were so important for early human survival. For our JONES surname things [genetically] took roots both in culture and language. Expanding their horizons they moved about the Iberian peninsula [and other places] and finally found their way up St. George's channel to place their Y-chromosome among the islands. For R1b1a - haplogroup [roughly 75% of us with the JONES surname ] the Welsh (Anglesey) at 89%, and Basque area (French, Spanish) at 88%, and the Turkic people (Bashkirs) were found with 86%. [confines of S.Ural mountains]
For those interested: 1) Irish = 82% , 2) Scots = 77%, 3) Spanish (Minorca) = 73% , 4) Dutch (Germanic west) = 70%. What a deal...that same Y-chromosome is found among most of those with the JONES surname of Welsh descent.
Subscribe to:
Posts (Atom)